The transformation is the unexpected result of experiments on a genetic circuit the creatures use to discern friend from foe.
Researchers have studied Myxococcus xanthus for decades as a model system for social cooperation and bacterial gene regulation. While studying M. xanthus, mutants that overexpress two proteins the cells use to recognize close relatives, researchers discovered a previously unreported behavior: self-organization into circles a millimeter or more in diameter.
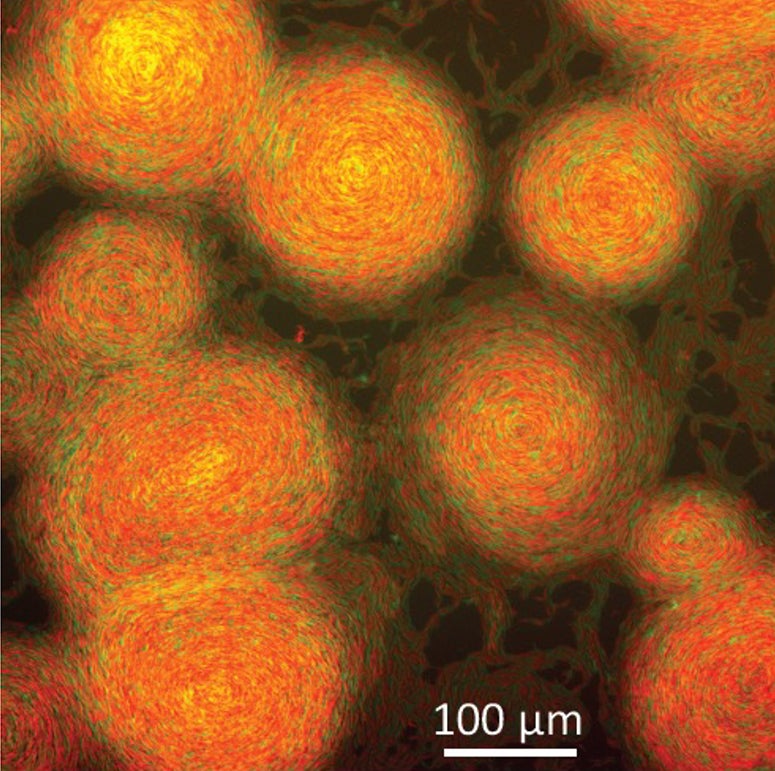
“When you overexpress that protein, you can see these circular aggregates emerge after four hours, and by 12 hours they take up the whole (petri dish),” says study coauthor Oleg Igoshin, a professor of bioengineering at Rice University and senior scientist at Rice’s Center for Theoretical Biological Physics.
Igoshin’s research group and a microbiology group from the University of Wyoming, led by coauthor Daniel Wall, collaborated for five years on the study, conducting dozens of laboratory and computational experiments to uncover the genetic mechanism of the circular swarms.
BACTERIA PREDATORS
M. xanthus prey on other bacteria. Lacking internal organs to digest their prey, they band together in familial packs to engulf and devour victims, which can include M. xanthus that aren’t members of the family.
About five years ago, Wall and coauthor Pengbo Cao, then a graduate student in his lab and now a postdoctoral research associate at the Georgia Institute of Technology, showed M. xanthus uses a surface receptor called TraA and a partner protein called TraB to recognize kin.
When M. xanthus bumps into a close relative, the TraAB complex acts as a kind of glue, forming a sticky bond between the two. When M. xanthus runs into unrelated M. xanthus, TraAB helps poison non-kin.
While investigating the mechanism of TraAB, Wall’s team created several mutant strains, including some that overexpressed TraAB, making more of the protein than normal. Cao noticed they had a tendency to form cell clusters within a few hours. While Wall’s team followed up with microbiological experiments, Igoshin’s group was asked if it could create a theoretical model that might explain the mystery.
“What’s interesting about our theory is that the only way we see these (circular aggregates) in our simulations is when we make the cells non-reversing,” Igoshin says. “In normal wild-type cells, they go back and forth, back and forth, like a commuter train. The head becomes the tail and the tail becomes the head. And they do it every eight minutes or so.”
The model simulated M. xanthus behavior based on changes to TraAB and other signaling circuits and was developed by Igoshin and study coauthors Rajesh Balagam and Zhaoyang Zhang, who were then graduate students in his lab.
“Our first idea was maybe they’re so sticky, they just can’t reverse,” Igoshin says. “So, we tried to see if sticky cells that normally reverse would form circular aggregates. We added a very, very strong adhesion to our simulations, and nothing happened. They didn’t make circles. However, if we instead inhibited the reversals the simulations worked. Circular aggregates emerged.”
EXTRA-STICKY MUTANTS
Follow-up experiments at the University of Wyoming verified cells in the aggregates did not reverse, but that raised even more questions.
“Somehow TraAB overexpression in aggregates prevented cells from reversing,” Igoshin says. “So this was very cool for us, because this is what our model predicted. But this was also a bit puzzling and completely unexpected. Because TraAB, as far as we knew, didn’t have anything to do with reversal regulation.”
The researchers ruled out a few possible explanations with follow-up experiments. Evidence suggested TraAB stickiness could be the key.
“But how is it that adhesion suppresses the reversals?” Igoshin says. “Our idea was maybe there is some sort of contact-dependent signal between cells that suppresses the reversals. The cells are in dense groups and are in contact with others all the time, but those contacts are transient. But if TraAB overexpression really makes you sticky, your neighbor will remain your neighbor for longer, and that could trigger the signal that suppresses the reversals.”
With those changes, the model began producing patterns very similar to what Wall’s team was seeing in its experiments with engineered M. xanthus strains.
To see if the model could predict a behavior that hadn’t yet been seen in experiments, Igoshin’s team simulated what would happen in mixed colonies of M. xanthus, including mixtures of two extra-sticky mutants that didn’t recognize one another as kin. The model predicted they would form large rotating swarms containing mixtures of the two strains.
Experiments from Wall’s team bore out the prediction, and the application of false color to microscopic images of the colonies revealed M. xanthus‘ pastiche of The Starry Night.
“Our work highlights how a social bacterium, known for rich sources of therapeutic natural products and as crop biocontrol agents, serves as a powerful model for studying emergent behaviors that also exhibit artistic beauty,” Wall says.
The paper appears in the journal mSystems. Additional researchers are from the University of Wyoming. The National Science Foundation and the National Institutes of Health supported the work.
[“source=futurity”]


